Data-Driven Clinical Planning: Building Value-Added Data Assets
With public data set to play an increasingly important role in drug discovery and development, an integrated, data-driven approach to clinical planning can optimize the probability of success.
The increasing availability of public data sets, such as The Cancer Genome Atlas (TCGA) and the Alzheimer’s Disease Neuroimaging Initiative (ADNI), offers a transformational opportunity for insight-driven planning at all phases of the drug development life cycle. The size and scope of these public data sets are often considerable, compiling billions of data points across multiple modalities and representing data that would have previously required many years and millions of dollars to generate. With unfettered access to these data, organizations can leverage this wealth of information to immediately adopt more rational approaches to trial design, patient stratification, repurposing, indication expansion, and portfolio strategy.
We see progressive organizations taking a strategic approach to compiling and integrating publicly available data to guide insight generation through a mixture of mechanistic and data-driven exploration. This data and knowledge asset can additionally be enriched with data from the organization’s own preclinical and clinical programs to exponentially increase an organization’s capability to develop and test hypotheses quickly and fuel a more data-driven approach to clinical planning.
As with any big data opportunity, one key to success lies in developing a systematic approach to harmonize, integrate, and analyze the data at hand. Too often, organizations are forced to employ a resource-intensive approach to data analysis that consists of just-in-time efforts to compile the data required to test a particular hypothesis or answer a program-specific scientific question as the need arises. This ad hoc approach represents a missed opportunity to instead build data assets that create both immediate and reusable downstream value through rapid and deep insight generation with compounding returns.
The opportunity within public data
Drug discovery and development have long been driven by the combination of data-driven and experimentally driven hypothesis generation and testing. The rapid growth of publicly available data increasingly affords an opportunity to accelerate this process of hypothesis generation and testing by taking advantage of existing data, with the goal of better targeting or reduced reliance on de novo lab experimentation.
At a minimum, this emphasis on data can advance a program more quickly and at reduced cost. At a more strategic level than financial and temporal optimization, we are seeing progressive biotechs seize opportunities to:
• Accelerate their path to the clinic by circumventing time-consuming, expensive data generation by leveraging data already available
• Increase the likelihood of success by mitigating the reproducibility risks inherent in preclinical models
• Quickly incorporate data-driven decision-making to guide program extensions and life-cycle planning
• Build a data asset that supports their eventual regulatory filings and articulates the value of their internal development programs
Example application
To demonstrate the opportunity presented by public data sets, QuartzBio, part of Precision for Medicine, extracted data from the lung adenocarcinoma data (LUAD) collection of TCGA. Using RNA sequencing data, significant changes in RNA expression were calculated among tumor adjacent tissue, tumor tissue, and between tumor tissues from patients classified as heavy or light smokers (>40 pack-year history vs <10 pack-year history). Reverse causal inferencing (RCI), a method that uses prior knowledge about a biological mechanism’s impact on gene expression to infer the activity state of that mechanism, was used to identify a stem cell-like molecular subtype associated with pack-year history of smoking. This stem cell-like phenotype was characterized by:
• Predicted decrease in the activity of pro-differentiation factors FOXP2 and PHOX2B
• Increased response to hypoxia, a feature of the tumor microenvironment with implications for de-differentiation of tumor cells, according to published literature[1]
COMPARISON
INFERED DIRECTION
BIOLOGICAL MECHANISM
ENRICHMENT
P VALUE
DIRECTIONALITY
P VALUE
All tumor vs
All adjacent
Decreased
mRNA expression of MIRLET7B
0.12
0.095 (not significant)
Increased
Response to hypoxia
0.18
1.1x10-8
Decreased
PHOX2B protein abundance
0.12
0.11
Decreased
Transcriptional activity of FOXP2
0.13
0.06
Increased
Transcriptional activity of TP63
1.8x10-2
3.2x10-3
Heavy tumor vs Light tumor
Decreased
mRNA expression of MIRLET7B
0.79 (not significant)
1.7x10-2
Decreased
Protein abundance of SOX6
0.14
3.2x10-3
Increased
mRNA expression of MIR140
2.5x10-2
0.11
Decreased
Transcriptional activity of HNF1A
7.0x10-2
7.6x10-3
Increased
Protein abundance of FAT1
6.3x10-2
0.11
Table. RCI to characterize LUAD pathogenesis. For this analysis, anything around or less than 0.1 was considered as a potential inferred mechanism describing the RNA expression data from the LUAD study. The enrichment P value evaluates the significance of the number of differentially expressed genes that were mapped to the biological mechanism network. The directionality P value evaluates the significance of the predicted direction (increased in green or decreased in red) of the network.
Analysis of patients with a more than 40 pack-year history of smoking compared with patients with a less than 10 pack-year history also suggested an augmented de-differentiation profile in those with higher cumulative inhaled smoke exposure. Therapeutically targeting tumors that exhibit a stem cell-like phenotype is an active area of research, and utilizing public data enables drug companies to determine whether the biology of the mechanism of action of their drug is represented in certain cohorts. One can imagine scaling this analysis systematically to prioritize indications that could potentially benefit.
Real-world uses
The aforementioned approach is one that we have seen biotech firms interested in addressing. Specifically, there is interest in:
• Building detailed mechanistic models describing the mode of action of their investigational drug
• Scanning public data in a systematic fashion to identify indications where the physiological mechanisms induced by the investigational drug could potentially ameliorate dysregulated disease biology
• Performing the analyses above on an individual patient, rather than at disease level, to identify those patients hypothesized to be most likely to benefit from the investigational drug’s mode of action
QuartzBio is assisting an organization in integrating all the information in TCGA to build a model using proprietary data on the investigational drug’s mechanism of action and its expected influence on human biology. The power of this model is that it may also identify potential noncancer indications with biological profiles that could be affected by the investigational drug, while also allowing for systematic identification of the best cell lines of preclinical models that recapitulate the disease biology represented by clinical samples.
Turning disjointed data into a true data asset
In order to analyze public data, the first step is to efficiently ingest, index, map, and globally harmonize diverse publicly available data sets so that relevant connections across types of biological (eg, genomic and transcriptomic) variation, across various data sources can be seamlessly made.
Of course, public data sets are incomplete for drug development. While we are fortunate to be operating in an era where the volume and variety of data generated across preclinical and clinical settings is increasing exponentially, such data are generated from a diversity of sources and typically remain disconnected due to their complexity and disparate nature. They remain siloed from publicly available data (Figure 1).
Figure 1. Data for clinical planning is siloed and disjointed
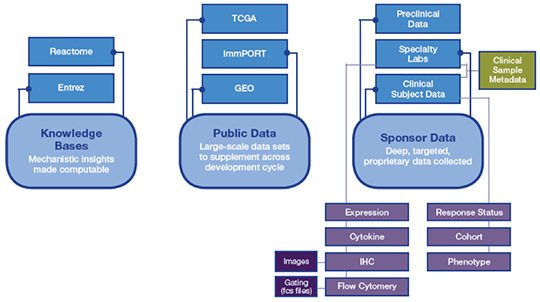
This represents a missed opportunity to enrich a sponsor’s proprietary data with data existing in the public domain or to enrich findings from any segment of data with biological understanding that exists in the literature.
Creating a data ecosystem
An ideal asset would integrate public data with proprietary sponsor data and link to scientific findings extracted from the literature as a computable knowledge base that allows contextualization of that data. For example, while identifying significant correlations among molecular entities in a data set is useful for generating hypotheses, it does not imply that there is a mechanistic, causal relationship between those entities. Conversely, a knowledge base can be used to rapidly determine whether other experimental findings demonstrated a similar correlation or even a causal relationship between the two. This strengthens the argument that a biologically meaningful relationship was identified and helps provide additional information about the function of that relationship in different contexts (Figure 2).
The result is a multidimensional, scalable approach toward generating actionable insights:
• Mechanistic approaches testing hypotheses derived from prior knowledge to attempt to explain the biological meaning behind a result. Examples of mechanistic approaches include reverse causal inferencing and gene set enrichment analysis (GSEA)
• Data-driven approaches identifying correlations in data and are generally amenable to any data type. These analyses can identify spurious relationships in the data, but often cannot explain biological mechanisms unless combined with prior knowledge. Examples of data-driven approaches include network reconstruction and weighted gene co-expression network analysis (WGCNA)
Figure 2. Schematic of a data ecosystem
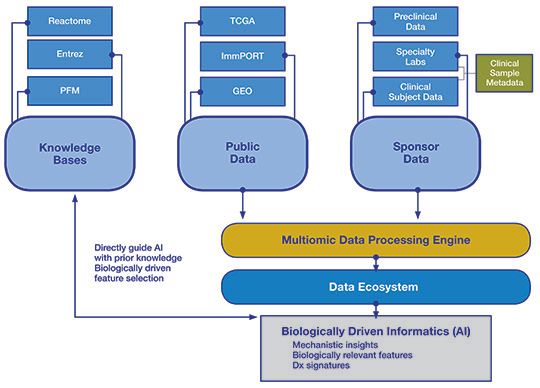
Leveraging a data ecosystem to unlock value
At an enterprise level, creating this type of integrated data ecosystem has a multitude of benefits across a portfolio, including the ability to:
• Capture information on why a drug does or does not work in certain patients. In studies where an end point is reached, sponsors experience a meaningful value inflection if they have a comprehensive data package that includes publicly available data. Even if an end point is not met, sponsors may still have clinically relevant data that enable them to adapt their programs
• Learn new information about the disease and investigational drug. These insights can be used to inform clinical development planning
• Validate the strength of evidence indicated by preclinical models. An increasing number of sponsors have become more and more skeptical about successful translation of preclinical models into the clinical setting. An integrated data ecosystem consisting of public data, knowledge bases, and sponsor data enables comprehensive evaluation of different preclinical models to identify the best fit. Using the abundance of disease clinical data available in the public domain, it is possible to evaluate different preclinical models to see where each model recapitulates human disease in a specific and mechanistic way
• Identify potential opportunities for combination treatments based on complementary or synergistic pathways
• Repurpose existing or previously abandoned compounds for new indications with stronger evidence
• Pivot programs more quickly based on real-time data and timely insight generation
• Build strategic data assets that not only add value to ongoing clinical trials, but also link back to drug discovery or patient selection
In short, implementing a systematic, scalable approach to support a combination of mechanistic and data-driven analyses enables organizations to make every bit of data matter.
Conclusion
In the years to come, public data will play an increasingly important role in drug discovery and development as these data are leveraged to enrich proprietary sponsor data and knowledge bases. This integrated, data-driven approach to clinical planning requires a thoughtful strategy and a multidisciplinary team with a cross-functional mix of big data and biological expertise. Ultimately, the investment in a data-driven mindset can increase by orders of magnitude an organization’s capability to advance the understanding of disease, discover new drug targets, identify biomarkers, stratify patients, and position drugs-ultimately streamlining the development process and optimizing the probability of success.
About the authors
Tobi Guennel, Ph.D, is Vice President and Chief Architect, and Renee Deehan-Kenney, Ph.D, is Vice President of Computational Biology, both at QuartzBio, part of Precision for Medicine.
[1] Kim Y, Lin Q, Glazer PM, Yun Z. Hypoxic tumor microenvironment and cancer cell differentiation. Curr Col Med. 2009;9(4):425-434.